

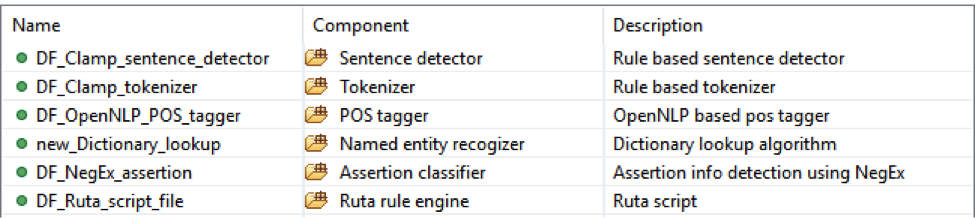
This is dictionary and rule based pipeline to extract smoking status from notes. It will recognize all smoking mentions and then category them into ‘NonSmoker’, ‘PastSmoker’ or ‘Smoker’;
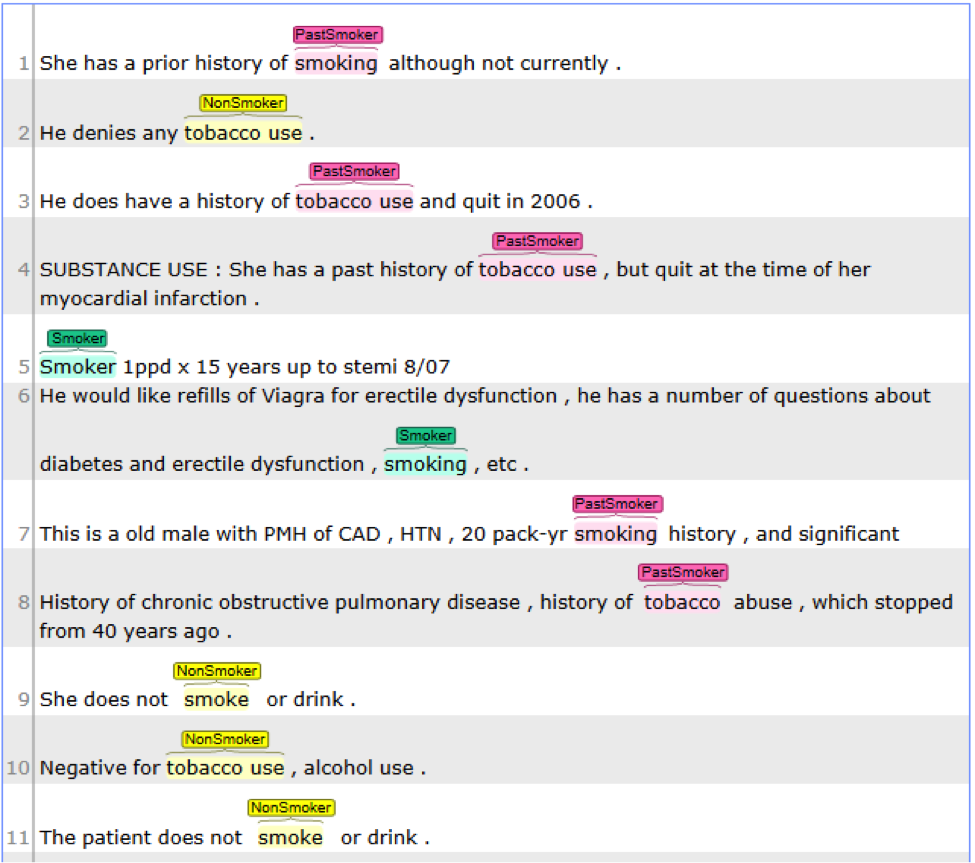
This pipeline will extract bleeding related concept and then map them to the UMLS CUI;
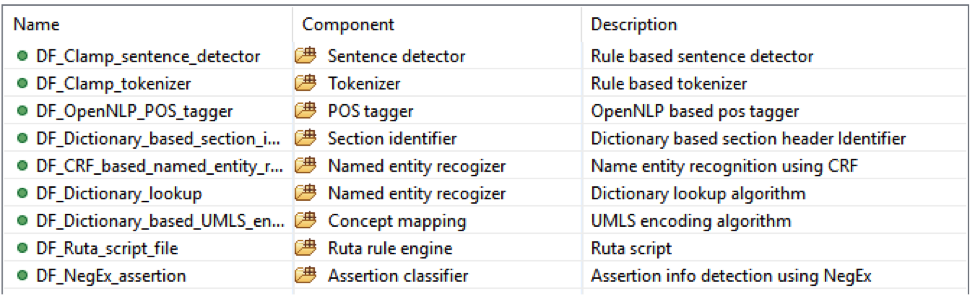
This pipeline will extract colorectal cancer mentions and then map them to the UMLS CUI;
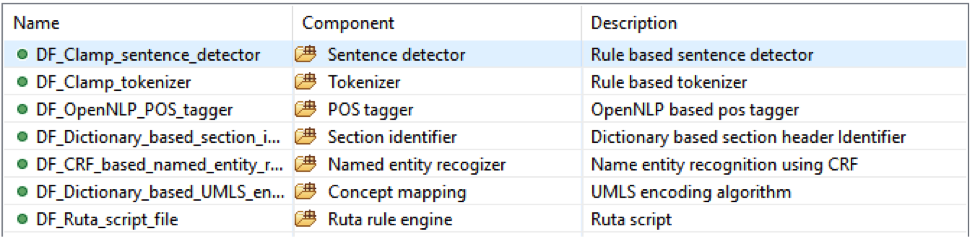
TThis pipeline will extract Site, Procedure, Histology etc. entities and relations among them from pathology notes.
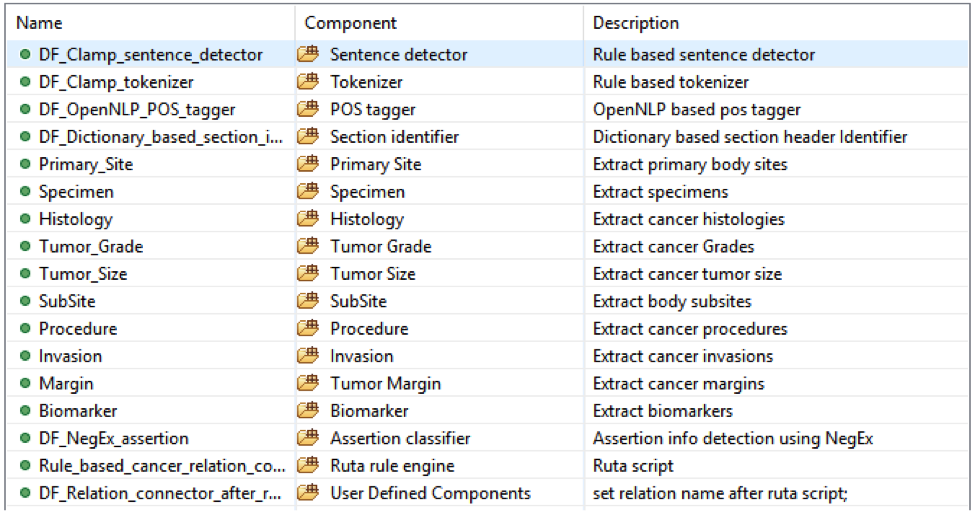
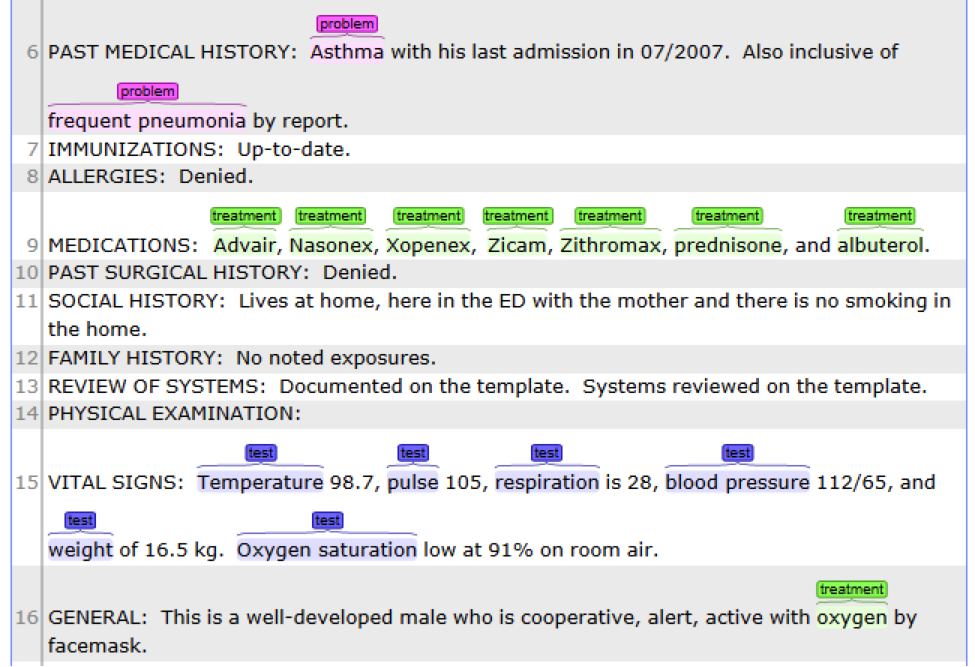
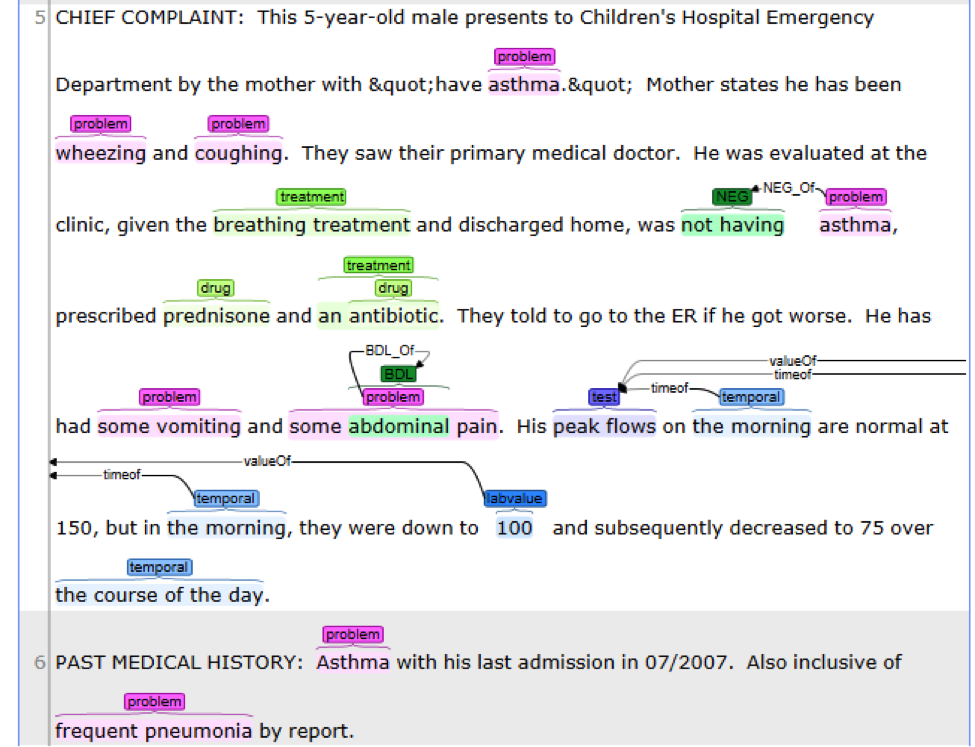
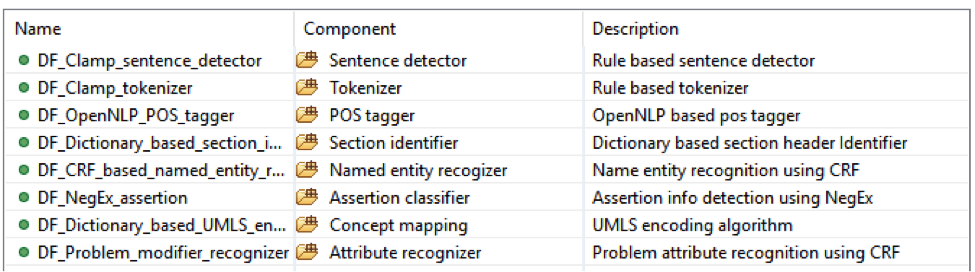
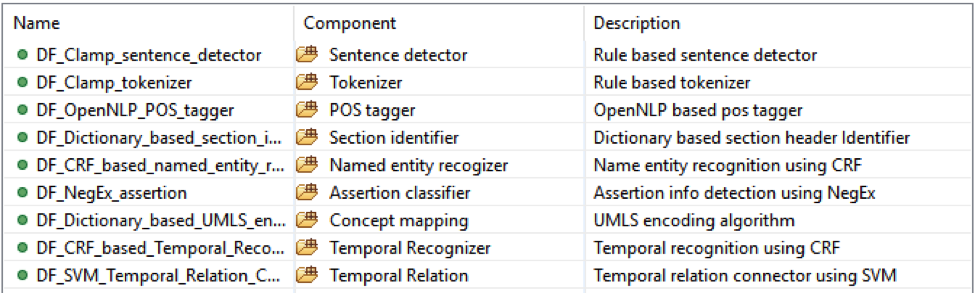
This is the CLAMP’s default named entity recognition pipeline. It will recognize ‘problem’, ‘treatment’ and ‘test’ from clinical notes and negation information of each concept. Users can add the UMLS encoder component to the end of this pipeline to get the UMLS CUI concept id (UMLS account is required for this component).
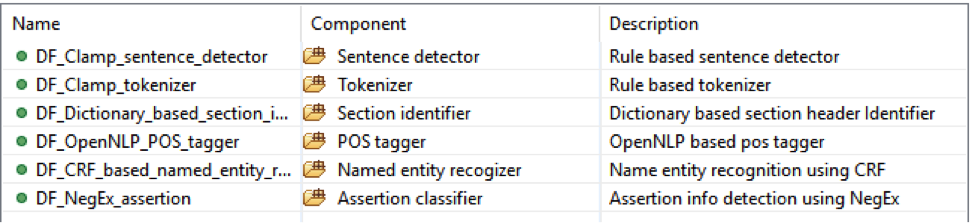
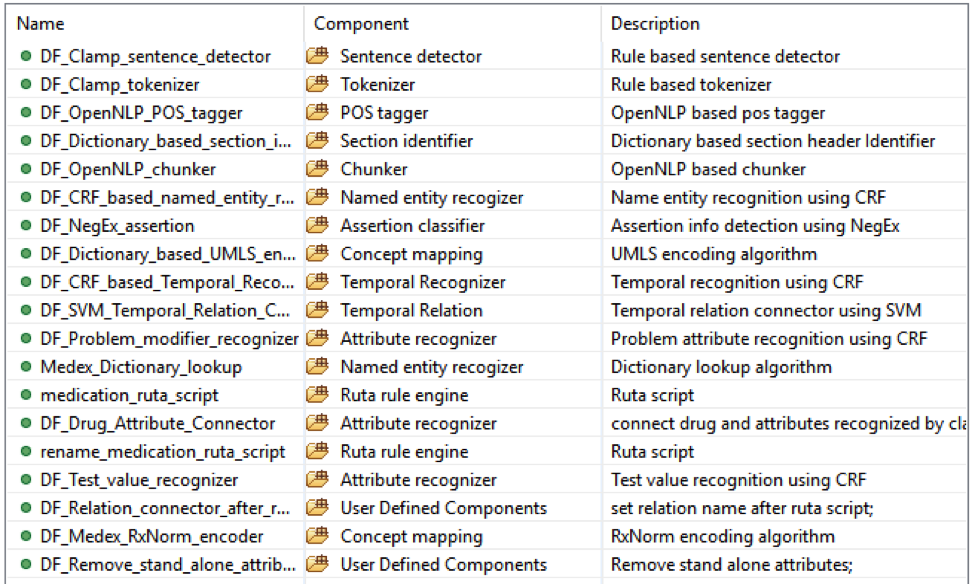
This is the default named entity recognition and relation extraction pipeline. The primary
entities include ‘problem’, ‘treatment’ and ‘test’. For each entity, the pipeline will recognize
its attribute as well.
• Problem with: subject, condition, negation, severity, location, uncertainty;
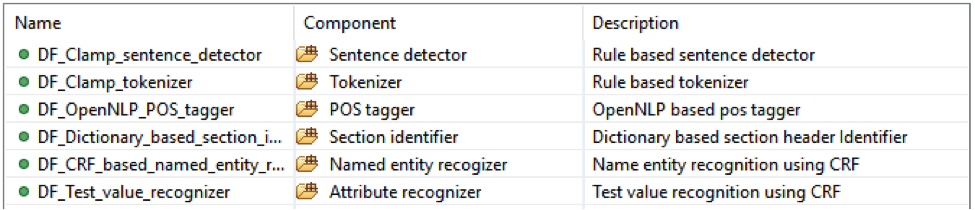
• Lab test with: test value;
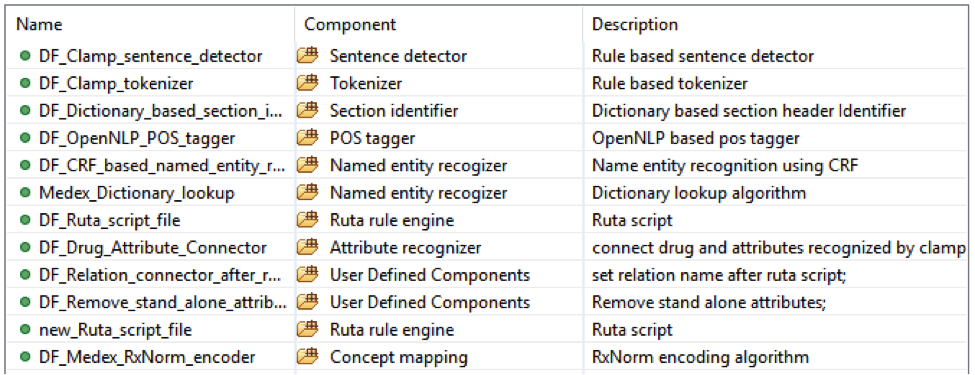
• Medicine with: dose, form, route, frequency, duration, necessity;
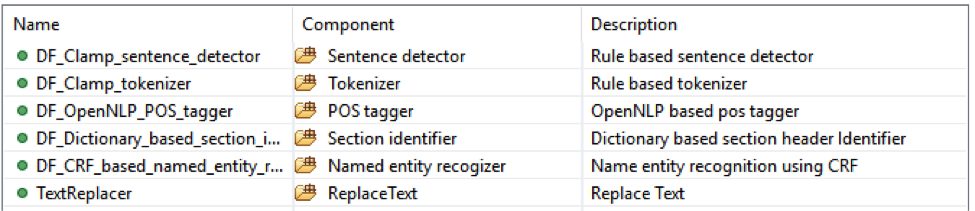
This is the de-identification pipeline. It will recognize all PHI information and then replace them with placeholder strings that are defined by the users. It contains 3 sub pipelines which are Disease-attribute, Lab-attribute and Medication-attribute.

Center for Computational Biomedicine
School of Biomedical Informatics
The University of Texas Health Science Center at Houston
7000 Fannin St, Houston, TX 77030
License Support
Hao Ding - Director of Operations
713-208-8195
Jianfu Li - Research Scientist
713-500-3934